|
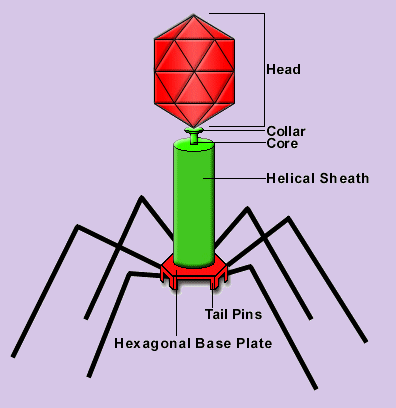
The T4 Virus
 A bacteriophage is a virus which infects bacteria. In particular, the
bacteriophage T4 is a virus which infects E.Coli, a bacteria that has been used
extensively for molecular biology research. The bacteriophage T4 exemplifies the life
cycle of viruses. It exists as an inactive virion until one of its extended 'legs' comes
into contact with the surface of an E. Coli. Sensors on the ends of its 'legs' recognize
binding sites on the surface of the host's cell, and this triggers the bacteriophage into
action. The bacteriophage binds to the surface of the host, punctures the cell with its
injection tube, and then injects its own genetic blueprint. This genetic information
subverts the host cell's normal operation and sets the cell's biosynthetic machinery to
work creating replicas of the virus. These newly created viruses escape from the cell and
then float about dormant until one happens to come into contact with a new host cell.
In nature, the bacteriophage T4 contains about 168,800 base pairs of
double stranded DNA. This genetic blueprint
contains all of the necessary information to create new bacteriophage T4. Our virtual
bacteriophage T4 also contains its own genetic information; it is made up of characters of
ascii VRML 2.0 code. At some point in the future evolution of cyberspace, our virus will
find an appropriate host and replicate itself. The virtual virus is born, and the question
of whether viruses are 'alive' is extended to the question of whether virtual viruses are
alive.
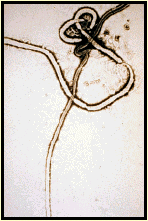
Ebola Virus
A Brief Epidemiological History of Ebola
The name "Ebola" is borrowed from a river in Zaire, Africa, where the virus
was first detected in 1976. That year there was also an outbreak in Sudan, resulting in a
combined total of about 600 cases, with a mortality rate between 53% and 88%, followed by
a significantly smaller Sudanese outbreak in 1979. In 1989, Ebola was detected in monkeys
imported from the Philippines to Reston, Virginia. Deadly to the monkeys, the Reston
strain did not afflict humans, although two people who had contact with the animals
produced Ebola antibodies. In fact, since 1979, not a single human case of Ebola virus
fever was identified for 15 years.
Then, in November 1994, a team of Swiss ethologists was in the Tai Forest of the Ivory
Coast to study a group of chimpanzees which had experienced two sudden bouts of mortality
over the space of a few years. In search of the cause, the scientists autopsied several of
the animals and discovered extensive hemorrhages. Following an autopsy on a chimp that had
died on November 16, a Swiss scientist came down with an atypical fever and was
hospitalized in Abidjan on November 26 with acute diarrhea and cutaneous eruptions. She
was evacuated to Europe, where she recovered.
As director of the Institut Pasteur's WHO Collaborating Center for Arboviruses and the
Viruses of Hemorrhagic Fevers, Professor Bernard Le Guenno conducted serological studies
of the Swiss ethologists and the chimpanzees. From a sample taken from the scientist
during the fever stage of her illness, Professor Le Guenno and his technician, Daniel
Coudrier, were able to isolate the virus by way of cell culture inoculations. Pierre
Gounon from Pasteur's Electron Microscopy Unit observed that it was an Ebola filovirus.
With the help of the CDC, it was further determined that it was antigenically different
from the other known Ebola strains. This strain, now named for the Ivory Coast, is likely
to have been responsible for the lethal epidemic observed among the chimpanzees and was
probably transmitted to the Swiss ethologist during her autopsy on the chimp. All other
people who had contact either with this woman or the animal's tissues tested negative for
the virus. This case provides the first documentation of monkey-to- human transmission in
the wild.
There was a resurgence of Ebola virus fever in Kikwit, Zaire, in 1995, traced to a
forester who fell ill in January of that year. Undiagnosed, he was treated at the Kikwit
hospital which in turn served as a transmission amplifier. Once the epidemic was
identified, a public awareness campaign about how to handle patients and the bodies of
victims helped to drastically reduce transmission. The last person fell ill on June 20,
1995. In all, 296 infected people were identified; only 20% survived. Cases of Ebola
infection have been identified as recently as last fall, when outbreaks in Gabon produced
approximately 100 cases causing about 60 deaths, the last on January 18, 1997. The
Gabonese goverment and WHO announced the official end of this epidemic in March. In an
article in The Lancet this past January, Professor Le Guenno established that a 1994
epidemic in Gabon had been misidentified as yellow fever and was in fact Ebola. The most
recent Gabonese outbreak led to the first case of Ebola fever in South Africa: a
Johannesburg nurse became infected after treating an undiagnosed Gabonese doctor for a
high fever last November. No other South African cases were reported.
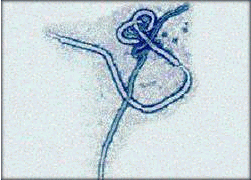
Preventing Future Epidemics
One suspected victim of the 1996 Gabonese out-break was transported to the Pasteur
Hospital in Paris, which for over 100 years has specialized in the treatment of infectious
diseases. The head of the hospital, Dr. Bertrand Dupont, describes the hospital's sanitary
system as one of extreme caution and vigilance where hospital personnel wear masks,
goggles, gloves, two gowns, etc., and pass through pressurized chambers before and after
contact with patients. All medical materials and equipment are disinfected and destroyed,
effectively killing the virus and preventing further transmission.
There is still no cure or treatment for Ebola virus fever. While it does not currently
threaten developed countries, a single undiagnosed case in any large Third World city
could potentially unleash several waves of infection with devastating consequences. This
scenario is unlikely thanks to the international network of WHO Collaborating Centers and
their rapid reponse to monitor and contain outbreaks. Where the virus is endemic,
education about handling the sick is essential and has proven very effective. Closer to
home, western hospital practices are likely to prevent any epidemic in the United States
or Europe. But until scientists identify the natural reservoir of Ebola, this virus will
remain a threat.

Professor Bernard Le Guenno in
Pasteur's highest-security-level
laboratory.
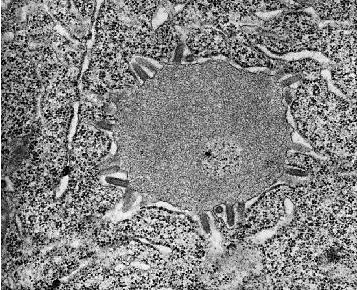
Electron micrograph of rabies virus in brain cells at 64,000 x
magnification. The bullets surrounding the smooth gray circle are rabies. The circle
itself is the Negri body, which can be seen with a light microscope. Courtesy of Dr. F.A.
Murphy, UC Davis
|
Mammals
Emerged Well Before Extinction Of Dinosaurs
Evidence from the largest evolutionary study of gene sequences ever performed shows that
the major groups of mammals emerged well before the extinction of the dinosaurs, according
to Penn State researchers.
Research by Sudhir Kumar, postdoctoral fellow, and S. Blair Hedges, associate professor of
biology, is published in the April 30 Nature.
"The evolution of mammals appears to have occurred gradually by the isolation of
breeding groups when the continents broke apart, not suddenly by the rapid filling of
ecological niches left vacant when the dinosaurs became extinct," Hedges says.
The massive gene study suggests that modern orders of mammals first evolved when the
continents were separating during the Cretaceous era about 100 million years ago -- much
earlier than some previous estimates based on fossil studies, which link the evolutionary
event to mass extinctions 65 million years ago.
"This is the first time we ever have been able to estimate when all these lifeforms
appeared on Earth," Hedges says. "Fossils can't give us this information, partly
because there are huge periods of Earth's history from which not enough fossils have been
found to make reliable estimates."
To gauge the pace of evolution, Kumar and Hedges mined a burgeoning collection of gene
sequences being accumulated at Genbank, the public genetic databases maintained by the
National Institutes of Health.
"During the past few years there has been a tremendous explosion in the number of
known gene sequences, so we had ten times as much data to work with as we did just two
years ago," says Hedges, who published a similar but much smaller study with Kumar
and others in 1996.
The scientists sifted through many thousands of vertebrate gene sequences from hundreds of
species to find those that develop mutations at a constant rate over time, which Kumar and
Hedges used like the ticking of a molecular clock to trace the history of each species
back to its origin.
Hedges says that, in contrast to the use of gene sequences, the use of fossils to estimate
when two species emerged from their last common ancestor
necessarily results in an under- estimation. "The body structure of a fossilized
animal had to have evolved at some earlier date before its lifetime -- in many cases, it
was many generations and many millions of years earlier. But genes are different -- their
clock-like mutations start ticking away as soon as a new species evolves, so the molecular
clock takes you back to the actual time of origin."
The researchers found that their molecular clock yielded origin dates similar to those
based on fossil dating for many species. But for others, the genetic clues lead back to a
much earlier time. For example, Hedges says "the fossil record for rodents says that
mouse and rat split only 10 million years ago, but we have 343 genes in this study telling
us that mouse and rat split from their last common ancestor 41 million years ago -- four
times as long. The rodent fossil record appears to have some major gaps," Hedges
says.
By comparing individual genes in pairs of species, the researchers identified 658 genes
from 207 vertebrate species that had accumulated mutations at a
fairly constant rate relative to one another during their evolution.
The scientists then calibrated this molecular clock to an evolutionary event well
established by fossil studies -- the divergence of birds and mammals about 310 million
years ago.
"A clock isn't any good unless it is calibrated to a time that everyone else agrees
on," Hedges explains, "and just about everyone agrees on the date when reptilian
ancestors of birds and mammals appeared because it is based on well-accepted studies of
fossils." Using this date as a secure calibration point -- and the mutation rate for
each of the constant-rate genes as a timing device -- the researchers were able to
determine how long ago each vertebrate order originated.
"We mined a lot of data by sorting through all the gene sequences in Genbank to find
the ones that fit our criteria," says Kumar. "The only way we could have
analyzed all the data so quickly was by taking a 'bioinformatic' approach," he says,
explaining that they used computers as well as manual methods to compare thousands of gene
sequences and test them for rate constancy using different parameters.
"It very easily is the largest evolutionary study of gene sequences ever done for
estimating the origin of so many species," Kumar comments.
Very few fossils resembling modern mammals or other vertebrates have been found in rocks
formed during the Cretaceous period, says Hedges, partly because paleontologists hardly
ever look for mammals in rocks that old.
"There has not been enough convincing evidence until now for paleontologists to
invest their time and money looking for mammal fossils in a time before
the dinosaurs became extinct," Hedges says. In addition, many scientists believe that
a large number of species suddenly sprang into existence at the very end of the Cretaceous
period.
Hedges says he hopes, as a result of this research, that paleontologists will now begin
searching for vertebrates in geological strata where they have never looked before.
"We are saying mammals definitely were living on Earth during the Cretaceous period
from 70 to 100 million years ago. We don't yet know what they look like, but from the
genes of their descendants we now know that they were there."
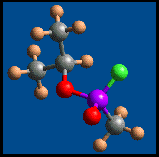
Sarin
Gas Molecule
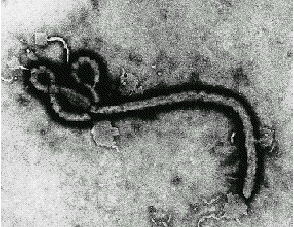
Filoviridae
Marburg Virus: etiological agent of an outbreak among lab monkey handlers in
Germany. In 1967, there was a simultaneous outbreak of HF in Marburg, Frankfurt, and
Belgrade among laboratory workers engaged in preparing kidney cell cultures from Africian
green monkeys. Additional cases involved attending medical staff. In all, there were 31
cases with 7 deaths.
Ebola Virus: causative agent of a severe, but short-lived outbreak in Africa.
Fatality rates may approach 90% with the Zaire strain of the virus. Outbreak in U.S in a
monkey quarantine facility in Reston, Virginia. The monkeys were sacrificed and the
facility was sterilized. No human cases were documented although four workers at the
facility sero converted.
These viruses are poorly characterized because their pathogenicity makes them difficult to
work with in the laboratory. They are classIV agents and can only be cultured in total
containment facilities. It is thought that they are enveloped viruses with negative-sense
genomes.
Ebola and Marburg Virus
"The head of a dress-maker's pin can provide seating accommodation for five
hundred million rhinoviruses (cause of the common cold) !"
Genome
|